Cas protein search
WHAT IS CasPEDIA?
CasPEDIA, or the Cas Protein Effector Database of Information and Assessment, is an encyclopedia of Class 2 CRISPR systems, presented in wiki format. Wiki entries provide comprehensive descriptions of enzyme activities, structures and sequences, complete with a literature review covering each nuclease's discovery, experimental considerations and applications.
CasPEDIA introduces CasIDs, which organize Cas enzymes by their functional properties. Inspired by the ENZYME Classification (E.C.) system, CasID nomenclature is composed of three digits, described below, that concisely describe effector properties and enable rapid identification of appropriate enzymes for a researcher's use case. CasIDs can be used to navigate the website through direct search in the search bar and using our Tool Finder.
Please visit the FAQ section for directions on navigating CasPEDIA and information about becoming a curator. Additional details about CasPEDIA are available in our preprint. Individuals interested in learning CRISPR fundamentals are encouraged to read CRISPRpedia, created by The Innovative Genomics Institute.
CITATION
CasPEDIA Database: A Functional Classification System for Class 2 CRISPR-Cas Enzymes
Adler, B. A.*, Trinidad, M. I.*, Bellieny-Rabelo, D., Zhang, E., Karp, H. M., Skopintsev, P., ... & Doudna, J. A. (2023). Nucleic Acids Research. doi: 10.1093/nar/gkad890
* Authors contributed equally
Cite with
EndNote or
BibTeX
DEFINING CasIDs
The three digits of CasID represent nuclease activity, target requirement, and guide RNA (gRNA) design, respectively. Definitions for all CasID digits are provided below.
To demonstrate CasIDs, consider SpyCas9a, with a CasID of 1.1.1.
- The Nuclease Activity for this enzyme falls under category 1, implying it has blunt double-strand cis nuclease-activity, and no trans nuclease-activity.1.
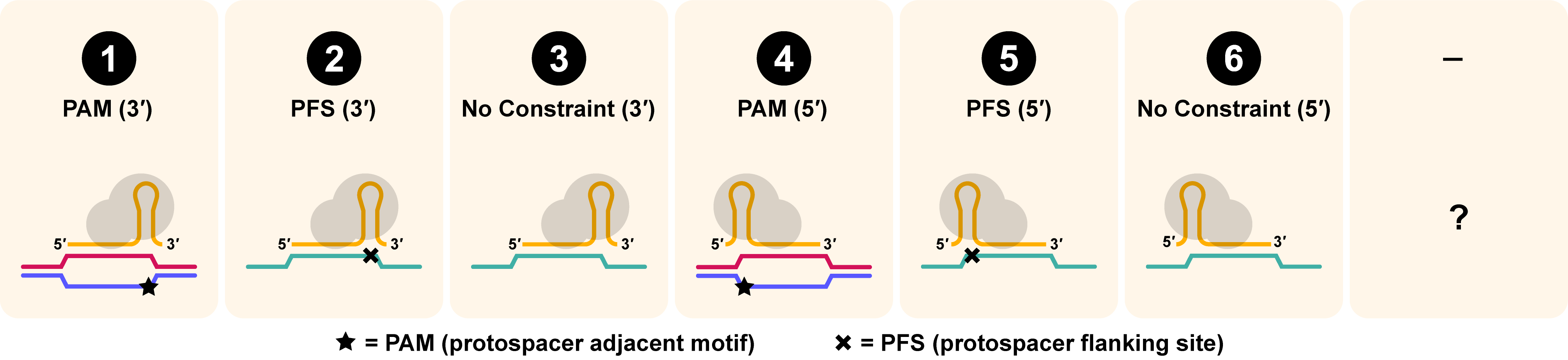
- The Targeting Requirements for SpyCas9a fit category 1, meaning it requires a 3' protospacer-adjacent motif (PAM)
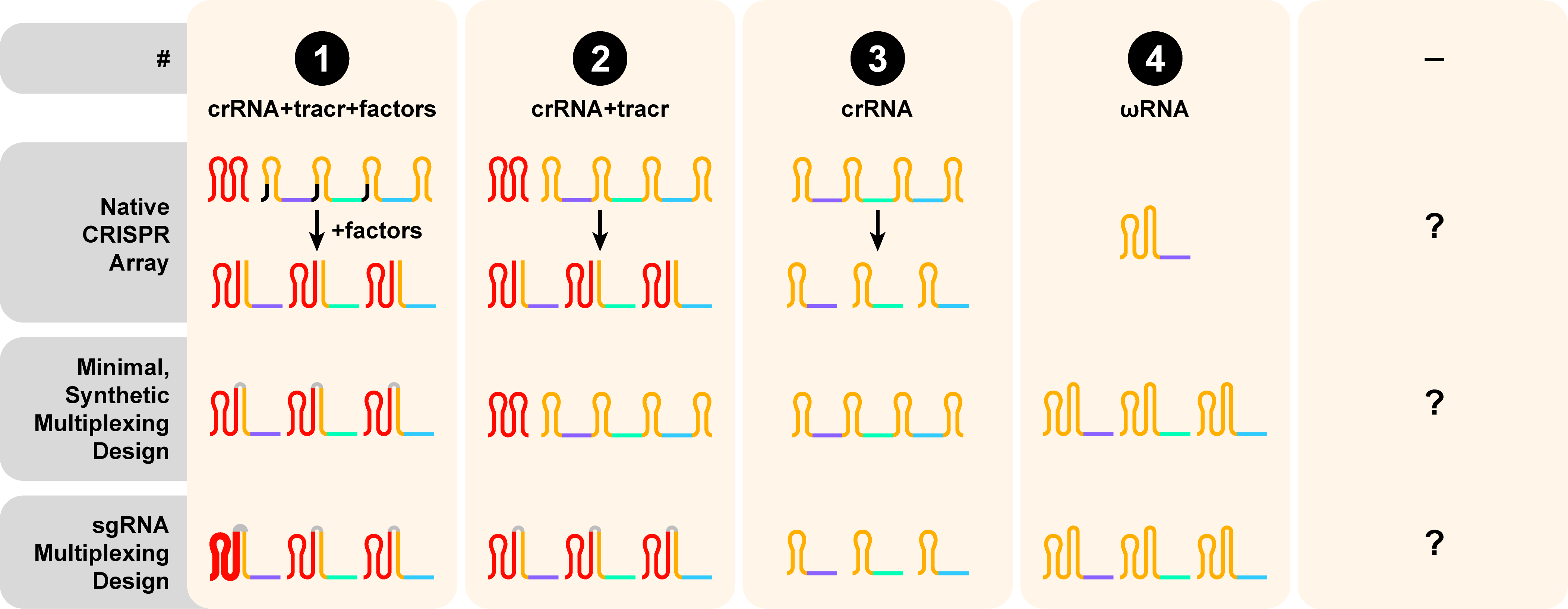
- Finally, the Guide RNA (gRNA) Design and Multiplexing properties for this enzyme fall under category 1, such that the native CRISPR array for SpyCas9a requires a CRISPR RNA (crRNA) for targeting + a trans-acting crRNA (tracrRNA) for multiplexing to the protein backbone of the effector + additional factors for processing the array into mature guides. SpyCas9a can also be engineered to utilize a minimal array containing different single-guide RNAs (sgRNAs), represented as a contiguous, all-in-one crRNA + tracrRNA sequence.2,3.

Primary Nuclease Activity
Nuclease activity is reported as the first digit in the CasID. Enzymes from the Class 2 Cas superfamily have diverse nuclease activities, both in an RNA-guided manner on a targeted sequence (in cis-) and sometimes on non-targeted nucleic acids upon binding (in trans-). The numerical annotation for nuclease activity relates to the known nucleic acid substrates for both the cis- and trans- activities of these enzymes. DNA sequences are shown in red and blue. RNA sequences (non-gRNA) are shown in green. Cleavage sites are shown with black triangles (▲▼). RNA guided Cas enzymes are shown in a fixed orientation to the nucleic acid substrate and not the value inferred by the Target Requirement.

Target Requirement
Target requirement is reported as the second digit in the CasID. Target requirement refers to the orientation of the Cas enzyme relative to its RNA-guided target substrate, as dictated by its protospacer adjacent motif (PAM), for DNA substrates, and protospacer flanking site (PFS), for RNA. Briefly, a PAM is an adjacent sequence required for targeting DNA and a PFS is a neighboring sequence that suppresses targeting activity. The numerical annotation shows the orientation of the PAM, or PFS, relative to the protospacer and dictates the orientation of the ribonucleoprotein (RNP) relative to its targeted substrate. DNA sequences are shown in red and blue. RNA sequences (non-gRNA) are shown in green. PAMs (required adjacent sequences) are shown with a star (★) and PFSs (excluded adjacent sequences) are shown with an X.
Guide RNA (gRNA) design + Multiplexing
Guide RNA (gRNA) design and multiplexing ability is reported as the third digit in the CasID. Guide RNA (gRNA) design illustrates how the properties of the native CRISPR array inform the creation of a synthetic gRNA scaffold. The numerical annotation describes the relevant guide requirements in several contexts: within the native CRISPR (top), the minimal synthetic guide design for multiplexing (middle), and the sgRNA design for multiplexing (bottom). Spacers are represented by purple, green, and blue lines. CRISPR repeats that are incorporated in mature gRNAs are colored in orange and black if they’re lost during crRNA processing. Trans-acting RNAs (tracrRNA) are colored in red. Synthetically-added sequences (i.e. tetraloops) are shown in gray.